Microfluidics — the phenomenon that underlies ‘lab-on-a-chip’ technologies — has long been of interest to medicine. Its ability to pass samples of fluids through channels containing compounds that bind to specific chemical groups or regions on chemicals in the sample make it potentially useful both in diagnosing diseases from body fluid samples taken from patients and in testing the activity of potential drug molecules. A team from the University of Michigan at Ann Arbor has now devised a microfluidic chip with another use — separating circulating cancer cells in blood samples.
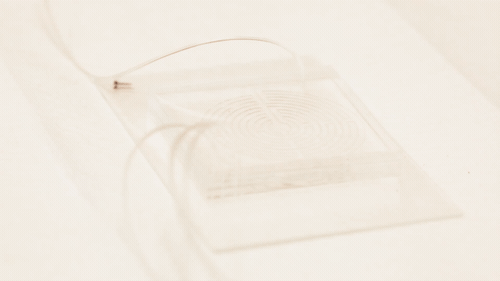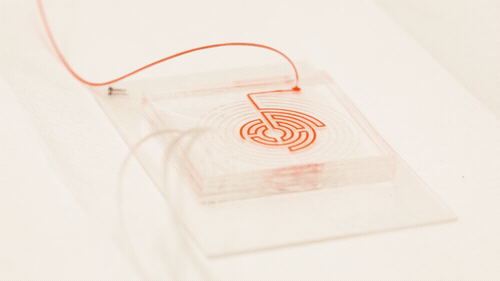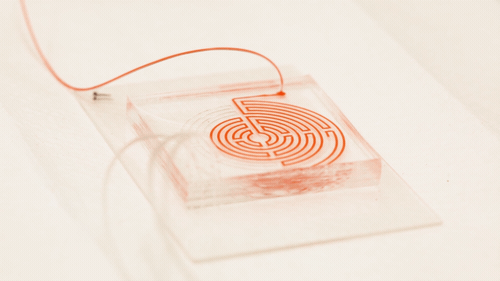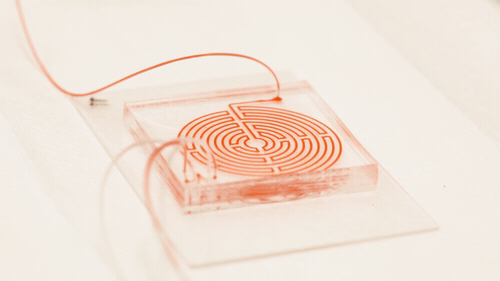
Circulating cancer cells break off from malignant tumours and roam through the body. They may latch on to other tissues and grow into new tumours, but they also might be important to treating the original cancer: if they could be isolated and studied, doctors could design a targeted drug therapy for the individual patient; monitor the cancer for genetic changes, and gain an early warning that the disease is about to spread. But the cells are elusive; they only account for a tiny proportion of blood cells, and they can transform into a type of drug-resistant stem cell while travelling through the bloodstream, making the usual method for targeting and grabbing cells — using enzymes that bind onto proteins on the cell surface — unreliable.
“You cannot put a box around these cells,” said Michigan chemical engineer Sunitha Nagrath, who co-led the microfluidic research along with oncologist Max Wicha, a pioneer in the use of cancer stem cells in therapy.
Microfluidics has been used in attempts at circulating cancer cell separation before, sending blood samples into a spiral-shaped channel etched into a chip. This was thought to be a good option because the cells are larger than healthy blood cells, but it proved to be imprecise; white blood cells tended to contaminate the separated cancer stem cell samples.
In a paper in the journal Cell Systems, Nagrath describes a chip featuring a much more tortuous and labyrinthine variation of the spiral path for the sample to take. Each time the path curves, eddies are generated in the flow which push particles in the sample away from the wall, with small particles pushed nearer the wall than larger ones. "Bigger cells, like most cancer cells, focus pretty fast due to the curvature. But the smaller the cell is, the longer it takes to get focused," Nagrath said. "The corners produce a mixing action that makes the smaller white blood cells come close to the equilibrium position much faster."
"Bigger cells, like most cancer cells, focus pretty fast due to the curvature. But the smaller the cell is, the longer it takes to get focused," Nagrath said. "The corners produce a mixing action that makes the smaller white blood cells come close to the equilibrium position much faster."
Nagrath’s chip packs a path six times longer than the conventional spiral into the same area, with many more corners. The number of white blood cells contaminating the resulting cancer stem cell sample was reduced tenfold from the spiral-chip method, and could be reduced further by running the sample through another labyrinth — a fast process, because there is no need to wait for the cells to bind to a targeting protein.
"We think that this may be a way to monitor patients in clinical trials," Wicha said. "Rather than just counting the cells, by capturing them, we can perform molecular analysis so know what we can target with treatments." He is already testing the device in a clinical trial with patients suffering from an aggressive form of breast cancer, trying to determine whether blocking interleukin-6, a protein involved in the normal healing process, could help prevent the circulating cells from changing into a stem-cell-like state, which is part of the mechanism of cancer spread.





Hard hat mounted air curtain adds layer of protection
Something similar was used by miners decades ago!